Gene Regulatory Neural Networks (GRNN)
Mobile App Design
A modern mobile app UI/UX designed for ASU Company focusing on user-centered interaction patterns.
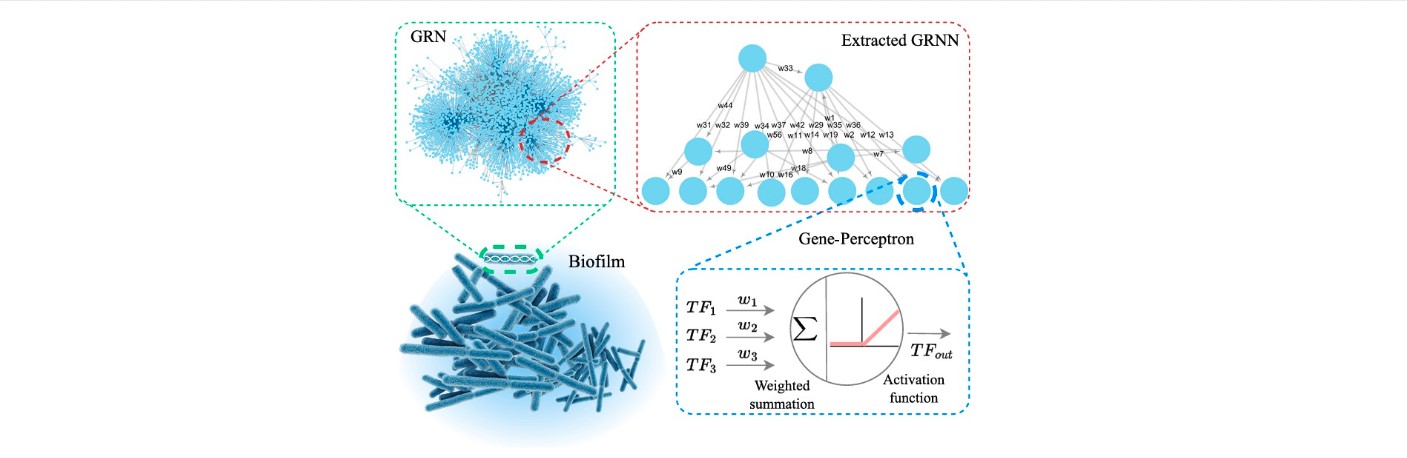
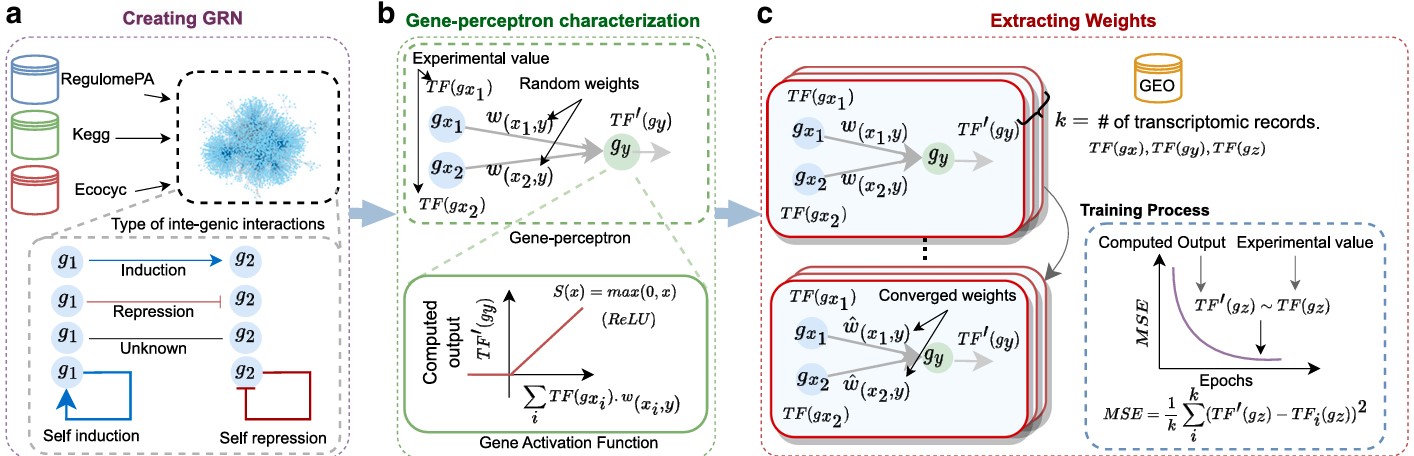
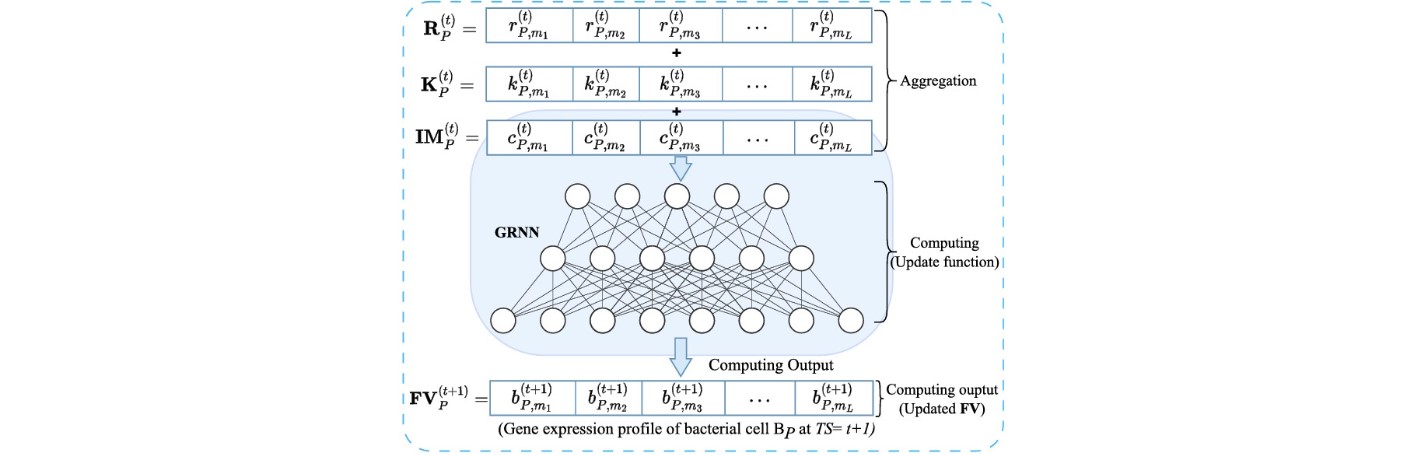
Gene Regulatory Neural Networks (GRNN)
We have introduced Gene Regulatory Neural Networks (GRNN) as a novel interpretation of bacterial gene
regulatory systems, mapping them into functional neural network models capable of computation within
living cells. This innovative framework enables biological systems to perform machine learning tasks by
leveraging the natural dynamics of gene expression and regulation.
Our GRNN architecture is designed to support wet-neuromorphic computing — a form of biocomputing where computation is carried out by living bacterial cells using gene-regulatory interactions. This approach not only offers adaptability and robustness but also drastically reduces energy consumption, making it highly suitable for low-power and edge AI applications.
Associated Publicatoins
- Somathilaka, S., Balasubramaniam, S., Martins, D. P., Li, X., “Revealing Gene Regulation-Based Neural Network Computing in Bacteria”, Biophysical Reports, vol. 3, no. 3, September 2023.
- Somathilaka, S., Balasubramaniam, S., Martins, D. P., “Analyzing Wet-Neuromorphic Computing Using Bacterial Gene Regulatory Neural Networks”, IEEE Transactions on Emerging Topics in Computing (Early access), March 2025.
- Somathilaka, Samitha, et al. "Wet tinyml: Chemical neural network using gene regulation and cell plasticity." Proceedings of TinyML Research Symposium, San Francisco, USA, April 2024.
Through this work, we are bridging synthetic biology and artificial intelligence, opening new pathways for sustainable and intelligent bio-hybrid systems.