Biocomputing
Mobile App Design
A modern mobile app UI/UX designed for ASU Company focusing on user-centered interaction patterns.
Biocomputing
Our research in biocomputing explores the use of Bacterial Gene Rgulatory Networks (GRNs) as intrinsic
computational systems. Rather than engineering synthetic circuits, we harness the native GRNN architecture
of bacteria such as E. coli to perform meaningful computation. Our key innovation lies in searching
for
application-specific sub-GRNNs within the natural network—enabling computation without genetic
modification.
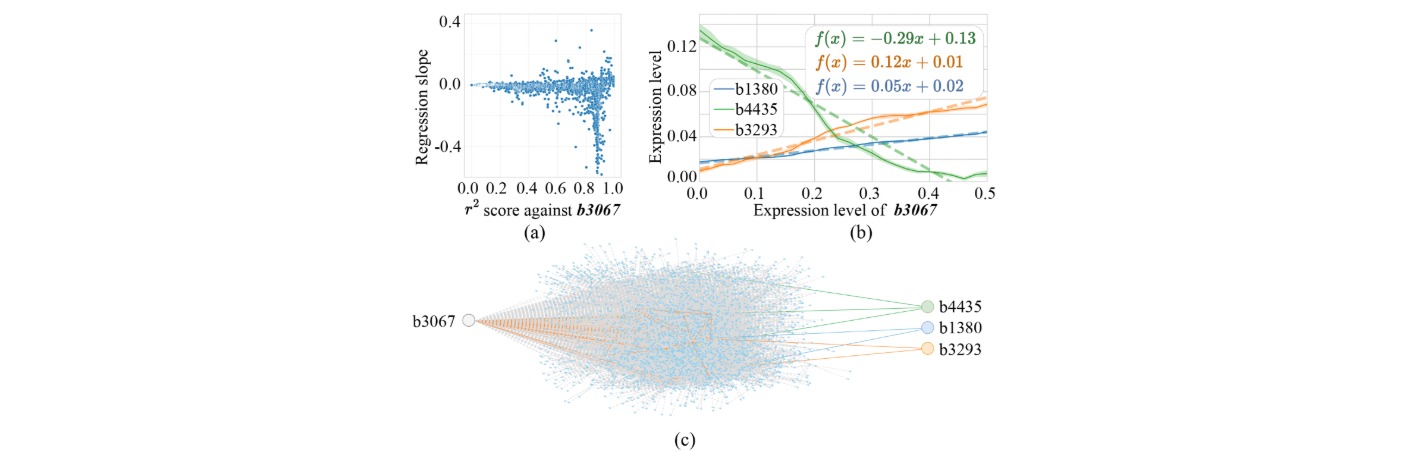
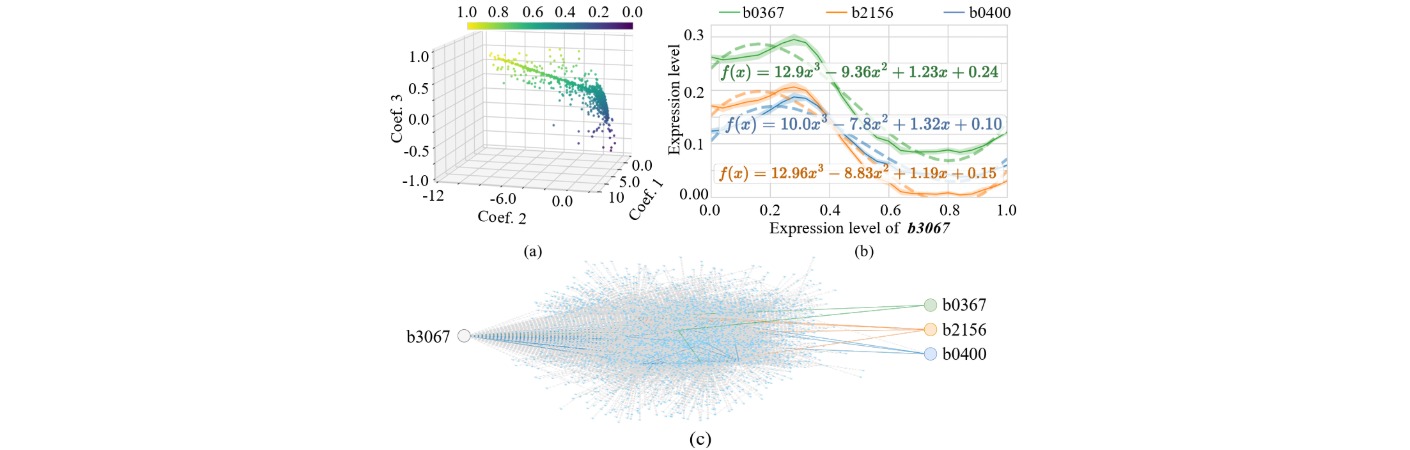
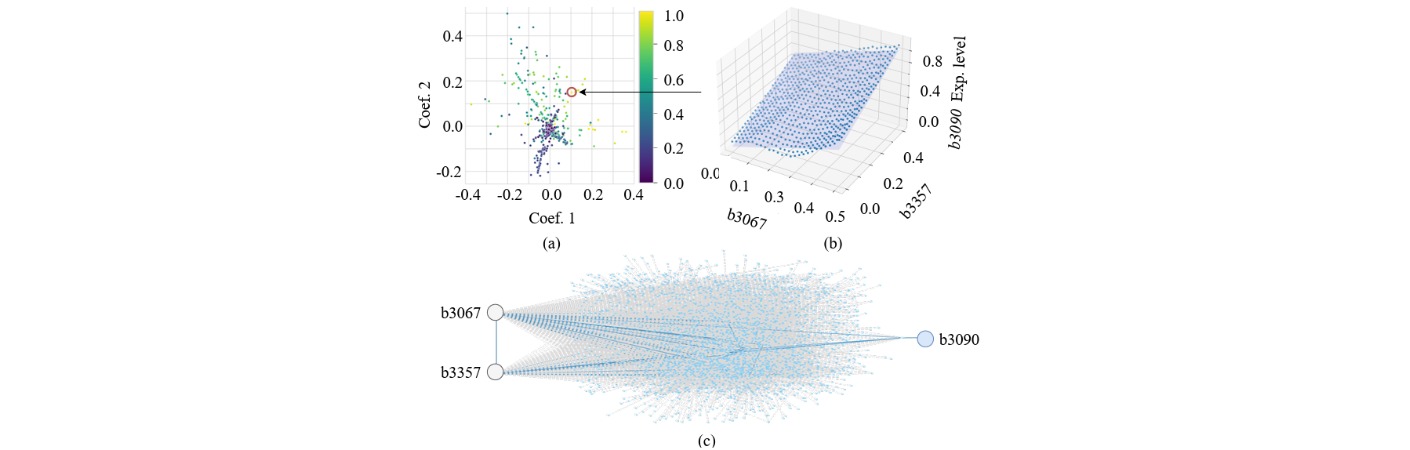
The inherent GRNN of E. coli can be viewed as a randomly structured neural network with approximately 5,000 nodes (genes) and over 10,000 regulatory edges, exhibiting diverse activation, inhibition, and combinatorial logic patterns. By identifying sub-networks that display computational potential, we extract regression- or classification-capable sub-GRNNs suited to specific tasks such as pattern recognition, signal transformation, or decision-making.
Associated Publicatoins
- Somathilaka, S., Balasubramaniam, S., Martins, D. P.,“Analyzing Wet-Neuromorphic Computing Using Bacterial Gene Regulatory Neural Networks”, IEEE Transactions on Emerging Topics in Computing (Early access), March 2025.
- Somathilaka, Samitha, et al."Wet tinyml: Chemical neural network using gene regulation and cell plasticity." Proceedings of TinyML Research Symposium, San Francisco, USA, April 2024.
- Ratwatte, A., Somathilaka, S., Balasubramaniam, S., Gilad, A. A.,“Non-Linear Classifiers for Wet-Neuromorphic Computing using Gene Regulatory Neural Network”, Biophysical Reports, vol. 4, no. 11, 2024.
- Ratwatte, A., Somathilaka, S., Balasubramaniam, S., Taggart, M., Nair, K.M., Niyato, D., O'Riordan, A. and Dooley, J., 2024."Reconfigurability of Gene Regulatory Neural Network for Regulating Biofilm Formation". IEEE Transactions on Network Science and Engineering, 2024.